Upload 22-05-2009 15:56 (Day-Month-Year, Paris time)
Update 28-09-2011 16:32 (Day-Month-Year, Paris time)
Information about the Author (who submitted the project in R.E.DD.B.)
Firstname Francois-Yves
Lastname Dupradeau
Institute UFR de pharmacie, UPJV
City Amiens
Country FRANCE
General information about the Project
Molecule keywords
| New amino acid |
O-methyl-L-tyrosine |
central fragment |
N-terminal fragment |
C-terminal fragment |
Abstract
RESP atomic charges embedded in force field libraries for the
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide dipeptide (or ACE-TYM-NME capped amino acid), as well as for the central, (+)NH3-terminal and (-)OOC-terminal fragments of the
O-methyl-L-tyrosine amino acid. All atom force field libraries suitable for MD simulations using the Duan
et al. AMBER force field.
Two conformations for
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide close to that found in the α-helix and/or β-sheet secondary structures were considered in the procedure. Charge derivation and force field library building for the central fragment of
O-methyl-
L-tyrosine (or TYM fragment) were carried out by using
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide, and setting two intra-molecular charge constraints to a value of zero for the ACE and NME residues during the charge fitting step. Charge derivation and force field library building for the
N-terminal fragment of
O-methyl-
L-tyrosine (or NTYM fragment) were performed by using two molecules: methylammonium and
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide, and setting two different constraints to a value of zero during the fitting step: (i) an inter-molecular charge constraint between the methyl group of methylammonium and the CH3CO-NH group of atoms of the capped amino acid, and (ii) an intra-molecular charge constraint for the NME residue of the capped amino acid. Charge derivation and force field library building for the
C-terminal fragment of
O-methyl-
L-tyrosine (or CTYM fragment) were carried out by using two molecules: acetate and
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide, and setting to a value of zero two different constraints during the fitting step: (i) an inter-molecular charge constraint between the methyl group of acetate and the CO-NHCH3 group of atoms of the capped amino acid, and (ii) an intra-molecular charge constraint for the ACE residue of the capped amino acid (Scheme 1).
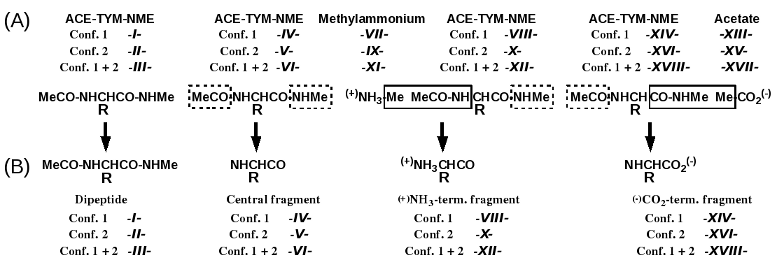 Scheme 1
Scheme 1
- Molecule -I-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix conformation) used in charge derivation & force field library building for the dipeptide itself.
- Molecule -II-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (β-sheet conformation) used in charge derivation & force field library building for the dipeptide itself.
- Molecule -III-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix & β-sheet conformations) used in charge derivation & force field library building for the dipeptide itself.
- Molecule -IV-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix conformation) used in charge derivation & force field library building for the TYM central fragment.
- Molecule -V-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (β-sheet conformation) used in charge derivation & force field library building for the TYM central fragment.
- Molecule -VI-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix & β-sheet conformations) used in charge derivation & force field library building for the TYM central fragment.
- Molecule -VII-: Methylammonium used in charge derivation & force field library building for the NTYM N-terminal fragment.
- Molecule -VIII-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix conformation) used in charge derivation & force field library building for the NTYM N-terminal fragment.
- Molecule -IX-: Methylammonium used in charge derivation & force field library building for the NTYM N-terminal fragment.
- Molecule -X-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (β-sheet conformation) used in charge derivation & force field library building for the NTYM N-terminal fragment..
- Molecule -XI-: Methylammonium used in charge derivation & force field library building for the NTYM N-terminal fragment.
- Molecule -XII-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix & β-sheet conformations) used in charge derivation & force field library building for the NTYM N-terminal fragment.
- Molecule -XIII-: Acetate used in charge derivation & force field library building for the CTYM C-terminal fragment.
- Molecule -XIV-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix conformation) used in charge derivation & force field library building for the CTYM C-terminal fragment.
- Molecule -XV-: Acetate used in charge derivation & force field library building for the CTYM C-terminal fragment.
- Molecule -XVI-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (β-sheet conformation) used in charge derivation & force field library building for the CTYM C-terminal fragment.
- Molecule -XVII-: Acetate used in charge derivation & force field library building for the CTYM C-terminal fragment.
- Molecule -XVIII-: N-Acetyl-O-methyl-L-tyrosine-N'-methylamide (α-helix & β-sheet conformations) used in charge derivation & force field library building for the CTYM C-terminal fragment.
Geometry optimization, frequency calculation and molecular electrostatic potential (MEP) computation were carried out using the Gaussian 03 program, while charge fitting was performed using the RESP program. For all the structures, the HF method and the 6-31G* basis set in gas phase were used in geometry optimization and frequency calculation. MEP computation was carried out using the B3LYP/cc-pVTZ theory level, the Polarized Continuum Model - the Integral Equation Formalism mimicking the diethylether environment, and the Connolly surface algorithm. The molecular orientation of each optimized structure was controlled using the rigid-body re-orientation algorithm implemented in the R.E.D. program. Four molecular orientations for
N-Acetyl-
O-methyl-
L-tyrosine-
N'-methylamide (based on the ACE carbonyl carbon, TYM carbonyl carbon, TYM carbonyl oxygen atoms; TYM carbonyl oxygen, TYM carbonyl carbon, ACE carbonyl carbon atoms; ACE carbonyl oxygen, TYM carbonyl oxygen, NME nitrogen atoms; NME nitrogen, TYM carbonyl oxygen, ACE carbonyl oxygen atoms), and two molecular orientations for methylammonium (based on the methyl carbon, nitrogen, gauche+ nitrogen hydrogen atoms; gauche+ nitrogen hydrogen, nitrogen, methyl carbon atoms) and acetate (based on the methyl carbon, carboxylate carbon, terminal oxygen atoms; terminal oxygen, carboxylate carbon, methyl carbon atoms)] were involved in MEP computation. A RRMS (relative root mean square value between the MEP values calculated by quantum mechanics, and those generated using the derived charge values) of 0.048 was obtained for the fitting step. See the F-23 up to F-44 R.E.DD.B. projects as well.
Publication YES 
Author(s) F.-Y. Dupradeau, A. Pigache, T. Zaffran, C. Savineau, R. Lelong, N. Grivel, D. Lelong, W. Rosanski and P. Cieplak
Journal Phys. Chem. Chem. Phys.
Year 2010
Volume 12
Page(s) 7821-7839
"Whole molecule" or "Molecule fragment" type project MOLECULE FRAGMENT
Interface R.E.D. used ? YES
Charge derivation procedure
Number of Tripos mol2 file(s) provided by the author(s) 12
Contain charge values & information about molecular topology
Information regarding Quantum Calculations
Geometry optimization
Program 1 GAUSSIAN 2003
Theory level 1 HF
More information 1 Opt=Tight
Basis set 1 6-31G*
Molecular electrostatic potential computation
Program 2 GAUSSIAN 2003
Theory level 2 DFT B3LYP
More information 2 IOp(6/33=2) SCRF(IEFPCM,Solvent=Ether)
Basis set 2 cc-pVTZ
Algorithm CONNOLLY SURFACE
Information about the charge fit
Program RESP
Number of stage(s) 2
input of stage 1 
input of stage 2 
Files the author of the project wishes to provide...
A script to convert Tripos mol2 file(s) into LEaP OFF library(ies) (for AMBER)... A script to convert Tripos mol2 file(s) into RTF or PSF library(ies) (for CHARMM)...
A script to convert Tripos mol2 file(s) into RTF or PSF library(ies) (for CHARMM)... A file to provide new force field parameters compatible with the Tripos mol2 file(s)...
A file to provide new force field parameters compatible with the Tripos mol2 file(s)... A file (choice made by the author) to provide more information about the project...
A file (choice made by the author) to provide more information about the project... A file (choice made by the author) to provide more information about the project...
A file (choice made by the author) to provide more information about the project... Download the whole project...
Download the whole project...



Internet page © 2006-2021. All rights reserved.
Université de Picardie Jules Verne - Sanford Burnham Prebys Medical Discovery Institute.
R.E.DD.B. projects free









