Upload 12-03-2009 18:15 (Day-Month-Year, Paris time)
Update 02-02-2011 09:42 (Day-Month-Year, Paris time)
Protection up to 12-03-2010 (Day-Month-Year, Paris time)
Information about the Author (who submitted the project in R.E.DD.B.)
Firstname Stephane
Lastname Abel
Institute CEA DSV/iBiTEC-S/SB2SM/LBMS
City Gif-sur-yvette
Country FRANCE
General information about the Project
Molecule keywords
| Alkyl glucosides |
Glucose |
Linear fatty alcohol |
Surfactants |
Membrane protein extraction/solubilisation |
Abstract
In the absence of force field topological fragments and RESP atomic charge values in the GLYCAM force field for linear alkyl glucoside surfactants used in membrane protein experiments, we have constructed a new force field topology database (FFTopDB) for alkyl α(1,4) and β(1,4) glucosides with long fatty chains [
i. e. -(CH2)n-CH3, with 5 < n > 11]. n-Octyl-β-D-glucopyranoside (OG), n-Dodecyl-β-D-maltoside (DDM), n-Undecyl-β-D-maltoside (UDM) and Decyl-β-D-maltoside (DM) are currently used in membrane protein extraction and solubilisation studies. [1]
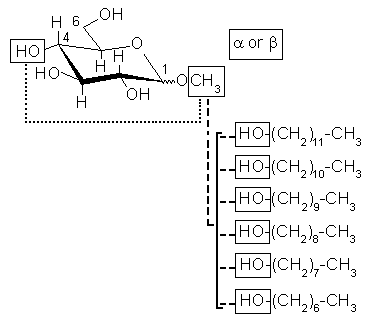
Scheme 1
Multiple molecules multiple conformations and multiple molecular orientations were used in charge derivation. Height molecules α-methyl-D-glucoside (AMG), β-methyl-D-glucoside (BMG), 1-heptanol, 1-octanol, 1-nonanol, 1-decanol, 1-undecanol, and 1-dodecanol were involved in charge derivation (Scheme 1). For the AMG and BMG molecules, two conformations were selected: ω dihedral angle (
i. e. O6-C6-C5-O5 and O6-C6-C5-C4) conformations
gauche,
gauche and
gauche,
trans were selected for the glucose. For the 1-heptanol, 1-octanol, 1-nonanol, 1-decanol, 1-undecanol, and 1-dodecanol alcohols the extended conformation was selected. Optimized geometries presenting intra-molecular hydrogen bond were excluded from the charge derivation. Thus, for glucose derivatives geometry optimization was carried out in presence of dihedral angle constraints. The HO4-O4-C4-H4 dihedral angle of the AMG and BMG molecules was constrained to 180°. Additional constraints for HO3-O3-C3-H3 and HO2-O2-C2-H2 were set to 180° for AMG and BMG, respectively. Frequencies were calculated for all the molecules, and transition state structures were excluded from the charge derivation procedure. Geometry optimization, frequency calculation and molecular electrostatic potential (MEP) computation were carried out using the Gaussian 03 program in gas phase, while charge fitting was performed using the RESP program. For all the molecules, the HF method and the 6-31G** basis set were used in geometry optimization and frequency calculation. MEP computation was carried out using the HF/6-31G* theory level, and the CHELPG algorithm. The molecular orientation of the optimized geometries was controlled using the rigid-body reorientation algorithm implemented in the R.E.D. program. Four molecular orientations for each glucose molecule (based on the C1 C3 C5, C5 C3 C1, C2 C4 O5 and O5 C4 C2 atom names) and for each alcohol (based on the C1 C2 C3, C3 C2 C1, C1 C2 C5 and C5 C2 C1 atom names) were generated before MEP computation, and involved in the charge fitting procedure to yield reproducible charge values. Charge fitting was performed using a single RESP stage, and a hyperbolic constraint value of 0.01. Glycolipide construction and connections between the different building blocks were carried by setting 14 inter-molecular charge constraints to a value of zero during the fitting step between the selected connecting groups: 12 inter-molecular charge constraints between the methyl group of α-methyl-D-glucoside or β-methyl-D-glucoside and the hydroxyl groups of 1-heptanol, 1-octanol, 1-nonanol, 1-decanol, 1-undecanol, or 1-dodecanol were used to build the acetal linkage of the glycolipids (dash line in Scheme 1). The construction of the α(1,4) and β(1,4) linkages between the glucose molecules was carried out by defining 2 inter-molecular charge constraints between the methyl group or HO4 hydroxyl group of α-methyl-D-glucoside and the HO4 hydroxyl group or methyl group of β-methyl-D-glucoside (ultra fine dash line in Scheme 1). The charge value of each hydrogen bound to a carbon presenting a sp3 hybridization was set to a constrained value of zero in order to insure a compatibility between this work and the GLYCAM force field. The charge derivation procedure was automatically carried out using the R.E.D. IV program. A RRMS (relative root mean square value between the MEP values calculated by quantum mechanics, and those generated using the derived charge values) of 0.148 was found in the fitting step.
[1] Raman, P. et al., The Membrane Protein Data Bank. Cellular and Molecular Life Sciences, 2006. 63(1): p. 36-51.
Publication YES 
Author(s) S. Abel, F.-Y. Dupradeau, E.P. Raman, A.D. Mackerell and M. Marchi
Journal J. Phys. Chem. B.
Year 2011
Volume 115
Page(s) 487-499
"Whole molecule" or "Molecule fragment" type project MOLECULE FRAGMENT
Interface R.E.D. used ? YES
Charge derivation procedure
Number of Tripos mol2 file(s) provided by the author(s) 10
Contain charge values & information about molecular topology
Information regarding Quantum Calculations
Geometry optimization
Program 1 GAUSSIAN 2003
Theory level 1 HF
More information 1 Opt=Tight
Basis set 1 6-31G**
Molecular electrostatic potential computation
Program 2 GAUSSIAN 2003
Theory level 2 HF
More information 2 IOp(6/33=2) NoSymm
Basis set 2 6-31G*
Algorithm CHELPG
Information about the charge fit
Program RESP
Number of stage(s) 1
input of stage 1 
Files the author of the project wishes to provide...
A script to convert Tripos mol2 file(s) into LEaP OFF library(ies) (for AMBER)... A script to convert Tripos mol2 file(s) into RTF or PSF library(ies) (for CHARMM)...
A script to convert Tripos mol2 file(s) into RTF or PSF library(ies) (for CHARMM)... A file to provide new force field parameters compatible with the Tripos mol2 file(s)...
A file to provide new force field parameters compatible with the Tripos mol2 file(s)... A file (choice made by the author) to provide more information about the project...
A file (choice made by the author) to provide more information about the project... A file (choice made by the author) to provide more information about the project...
A file (choice made by the author) to provide more information about the project... Download the whole project...
Download the whole project...



Internet page © 2006-2021. All rights reserved.
Université de Picardie Jules Verne - Sanford Burnham Prebys Medical Discovery Institute.
R.E.DD.B. projects free









